TrAC Trends in Analytical Chemistry Volume 152, July 2022, 116604,Text Full
Ting Wang【王婷】†, Yanlong Xing【邢艳珑】†*, Ziyi Cheng【程子译】, Fabiao Yu【于法标】*
Laboratory of Neurology, The First Affiliated Hospital of Hainan Medical University, Key Laboratory of Emergency and Trauma, Ministry of Education, College of Emergency and Trauma, Hainan Medical University, Haikou 571199, China
https://doi.org/10.1016/j.trac.2022.116604
Highlights
•A comprehensive review covering the recent progresses in single extracellular vesicle-based analysis.
•Discussion on the significance of single extracellular vesicle analysis.
•Focusing on the advanced analysis of single extracellular vesicles, especially in combination with microfluidic platform.
•Review on the single extracellular vesicle analysis-based biomedical application with emphasis on cancer investigations.
•Discussion on the existing obstacles and future perspectives on single extracellular vesicle research.
Abstract: Extracellular vesicles (EVs) are lipid membrane enclosed nano-sized vesicles that are secreted by all known organisms. These vesicles are increasingly recognized as important circulating biomarkers for the diagnosis and prognosis of different diseases including various types of cancer, owing to their essential role in intercellular communication. EVs preserve heterogeneity in both physical properties and cargos, which makes it extraordinarily tough to fully exploit their clinical potential. Therefore, comprehensive characterization of single EVs and their sensitive detection are urgently demanded. In this article, we survey the latest progress in single EVs analysis with innovative discoveries in heterogeneity and highlight the various label-free and labelling approaches of single EVs detection. Furthermore, the state-of-the-art advances in single EV-detection based biomedical applications with especial emphasis on cancer investigations are summarized. To the end, the challenges and prospects for exploiting new system in the field of single EVs study are discussed.
Introduction
EVs are heterogenous, lipid-bilayer-phospholipid membranous vesicles generated by various living cells through active secretion [1, 2]. Although initially thought to be cell debris, EVs have been discovered as vital biological species, owing to their physiological and pathological function in organisms. These vesicles carry bioactive molecules such as proteins and nucleic acids that are inherited from parental cells, and thus, affect microenvironment locally and at a distance by transferring cargos to recipient cells [3]. EVs can mediate intercellular communication and have been regarded as potential biomarkers for the diagnosis and treatment of diseases [4, 5].
EVs can be released by cells to the extracellular space via different ways. Based on the currently known origin mechanism of EVs, these vesicles can be divided into three categories: exosomes (30-200 nm in diameter), microvesicles (100-1000 nm in diameter) and apoptotic bodies (500-2000 nm in diameter) [2]. In this review, we concern on exosomes and microvesicles and collectively define them as EVs. Exosomes and microvesicles have different modes of biogenesis. In one aspect, exosomes are originated from endocytic pathway. Initially, inward budding of cellular plasma membrane results in the formation of early endosome. Further inward invagination and budding of membrane inside early endosome leads to the formation of multivesicular body (MVB) bearing intraluminal vesicles that carry transmembrane, cytosolic contents, and peripheral proteins. MVBs may then partially fuse with lysosomes and degrades inside cell. Alternatively, MVBs can fuse with plasma membrane and release vesicles to the extracellular environment, which are defined as exosomes. In another aspect, the direct outward budding of the plasma membrane induces the formation of microvesicles [6, 7]. Therefore, EVs preserve high heterogeneity in physical characteristics (size, density, morphology) and cargos (protein, lipid content, nucleic acids), mainly owing to their intricate biogenesis processes [8].
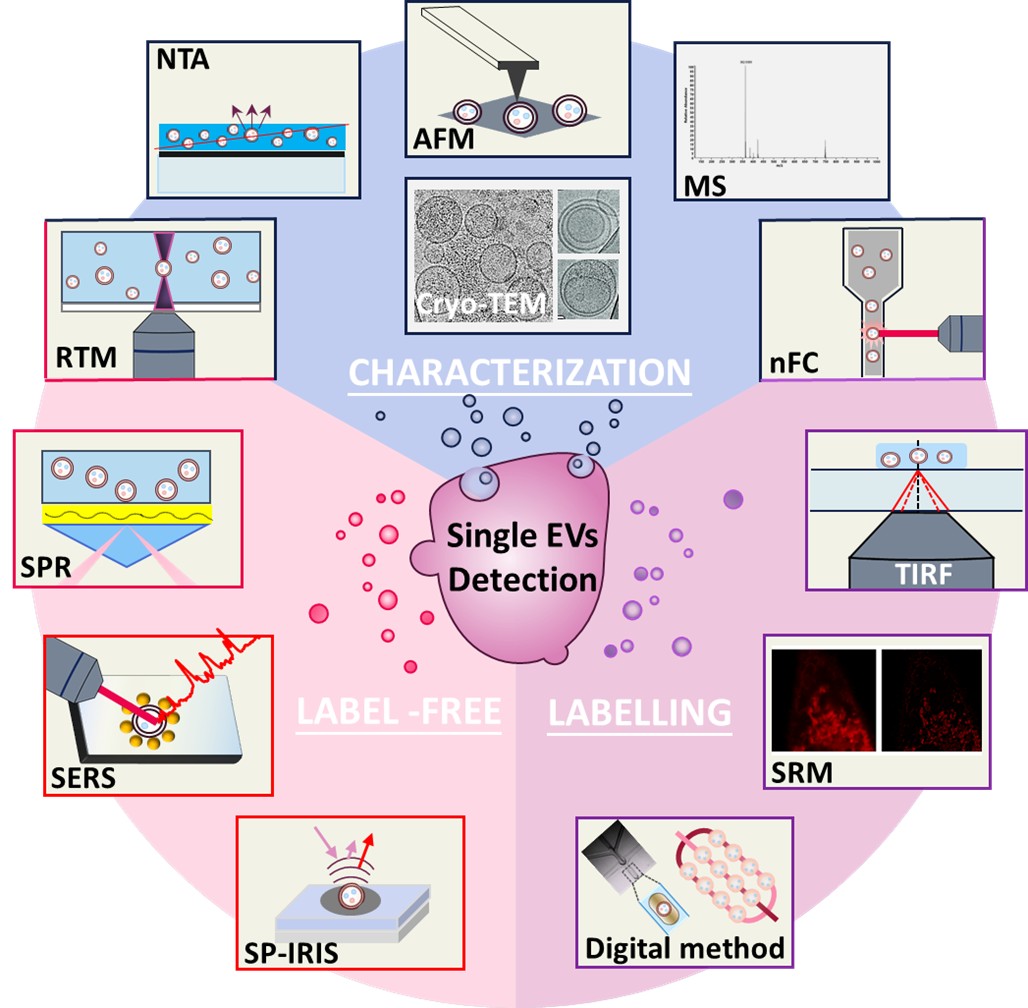
Growing evidence has demonstrated the role of EVs in the development of various diseases such as neurodegenerative diseases, acute organ injury and cancer, owing to the bioactive cargos carried and transferred by EVs [5]. In particular, tumour secreted EVs effect critical functions in facilitating intercellular communication in tumour microenvironment and modulate tumour initiation and progression [9]. Additionally, EVs are widely present in various bodily fluids and have advantages in high concentration (up to 1011/mL) and stability in the blood circulation. As a result, tumour derived EVs can be used as promising biomarkers for liquid biopsy in cancer patients [4, 10]. Notably, many tumour-associated protein biomarkers have been identified in EVs from clinical blood samples, and their types and expression levels are strongly correlated with the presence and progression of certain cancer, which makes the investigation on EVs’ heterogeneity important [11]. Since EVs are heterogeneous in sizes and contents, it is of utmost importance to investigate the molecular composition of EVs at single vesicle level in order to completely understand the biological function of various EV subtypes in disease development and exploit their clinical value [11]. Conventional techniques such as western blotting (WB), enzyme linked immunosorbent assay (ELISA) and real-time polymerase chain reaction (RT-PCR) have been utilized to detect EVs contents (protein and mRNA), however, mainly for bulk vesicles, which are unsuitable for single EV analysis [2, 5]. Recent years, research progress has been made in analysis of single EV, using various advanced detection technologies [12, 13]. In general, detection approaches of single EVs fall into two main categories, label-free and label-based techniques. Label-free methods such as Raman spectroscopy and plasmon resonance are based on the physical properties of EVs, offering non-destructive approaches for single EV detection [14]. Leveraging fluorescently labelled antibody or aptamer, label-based methods can achieve the detection of individual EVs, in conjugation with flow cytometry or fluorescence imaging techniques etc. [15]. Herein, we intend to summarize the recent progress in single EVs analysis and the latest advances in biomedical applications, with especial emphasis on cancer diagnostics based on EV-derived biomarker discovery. In detail, the influence of EVs’ heterogeneity on their function and the significance of single EVs analysis are discussed. Additionally, various characterization and detection methods are listed and compared. And the state-of-the-art clinical application in cancer diagnosis based on single EVs analysis are exemplified. Finally, we propose the current challenges and future perspectives in single EV-related research.
Significance of single EVs analysis
Cells actively release a large number of EV populations with distinct biomechanical properties and biological functions into the extracellular environment, which exerts diverse biological effects on recipient cells [16]. Exosomes and microvesicles are most concerned EV populations, which are discriminated based primarily on their cellular origin [3]. Nonetheless, increasing evidence has indicated that these EV populations contain various subpopulations with unique function in bioprocesses. For instance, non-membranous nanovesicles and exosome subsets including small and large exosome vesicles from various cancer cell lines have been identified [17]. Additionally, in a recent study, it has been observed that tetraspanins are unevenly distributed across single EVs [18]. The EV subpopulations can reflect the associated biological processes, which enables them to be promising biomarkers for clinical diagnosis. However, these intrinsic diversity and heterogeneity of EVs make it more complex and difficult for investigating their biology and function. Typically, in cancer biology, the various EV subtypes may have unique biological roles in the development of cancer, and the varied distribution of membrane proteins on single EVs may bias sensitivity to multiplexed cancer biomarkers [18, 19].
Therefore, one of the major concerns in EV research is to address the heterogeneity within EV populations and investigate the molecular composition of single EVs in detail [12]. Clarifying EVs’ diversity will help to better understand the exact function of EV in the physiological and pathophysiological processes, and ultimately accelerate the use of EVs in diagnostics and therapeutics. The significant progress has been made in the field of EVs, owing to the improved characterization and detection techniques, which enable deciphering of the heterogeneity of single EVs for biomedical applications [20].
Characterization techniques of single EVs
The characterization of EVs include two parts, one is the physical characterization including morphology, size and distribution, the other is the characterization of the molecular composition [21]. The physical characterization can only be employed to examine the physical characteristics of EVs, while complementary techniques are needed to examine the molecular composition e.g., protein, nucleic acids etc. to ensure the successful isolation of the desired vesicles [22]. There are multiple methods for the physical characterization of EVs including scanning electron microscopy (SEM), transmission electron microscopy (TEM) and dynamic light scattering (DLS) [5]. Different from the above techniques that are commonly employed for bulk EVs examination, as illustrated in Fig. 1, nanoparticle tracking analysis, atomic force microscopy and cryogenic transmission electron microscopy have been proved to be valuable methods for characterizing single EVs [23, 24]. Characterization of the molecular composition of single EVs requires optical techniques such as Raman or fluorescence spectroscopy to obtain the molecular features of individual vesicles, either with or without exogenous labels. Commonly, Raman tweezers microspectroscopy and nanoscale flow cytometry have been employed for the molecular characterization of single EVs [25, 26]. Mass spectrometry has also been employed to characterize EV proteins specifically expressed by single EV subpopulations [27]. The description and comparison of different characterization techniques are summarized in Table 1.
Table 1. Summary of characterization techniques for single EVs
| Techniques | Working principle | Information obtained | Advantages | Disadvantage | Ref |
| Physical characterization (morphologies, size and distribution) | Nanoparticle tracking analysis (NTA) | Tracking and recording the Brownian motion of nanoparticles in suspension | Size distribution and concentration of particles | Minimum sample preparation, easy to operate, less time-consuming | Low specificity, possible false signal from protein aggregates or other nanoparticles, interference of scattered light from adjacent particles | [5, 32] |
| Atomic force microscopy (AFM) | Measuring the interaction between the probing tip and sample surface | Morphological and mechanical characteristics of single EVs | No special sample preparation, high resolution, high throughput, integration with other techniques | Limited information obtained; perfectly flat substrate required, time consuming, labour intensive | [28, 29] |
| Cryogenic transmission electron microscopy (cryo-TEM) | Observing frozen samples under transmission electron microscopic | Morphology and structure | No complicated sample preparation, retain the native structure of EVs, high resolution | Limited information obtained, low throughput, low contrast image | [31, 33] |
| Molecular composition characterization | Raman tweezers microspectroscopy (RTM) | Measuring the Raman spectra of vesicles using laser mediated tweezing of the object | Raman fingerprints of the sample EV’s chemical constituents | No exogenous label, non-destructive, timesaving, no sample treatment | Weak signal, low throughput | [25, 34] |
| Mass spectrometry | Measuring mass-to-charge ratio after ionization and fragmentation of sample molecule in the gas phase | Molecular mass and ion fragmentation pattern of given molecule showing the structural information | No exogenous label, rapid, sensitive, specific, high throughput | Limited information obtained, strict operation condition, coupling with other techniques required | [27, 35] |
| Nanoscale flow cytometry (nFC) | Detecting the multiparametric scattered light and fluorescence signal emitted by labeled vesicles on a nanoscale flow cytometer | The size distribution and diversity of EV populations, and the protein or nucleic acid content of single EVs | High resolution, high throughput | Interference from unbound dye, highly purified labelled EVs and properly diluted samples required | [26, 36] |
| Multiparametric characterization | AFM-IR, NTA-TIRF | Different techniques are coupled on one setup to measure the same sample simultaneously and measure by two techniques | membrane protein composition and abundance, the size and mechanical properties etc. | Multiparametric analysis of single EVs, detailed information on morphology and structure | Special setup required, different software for processing and analysis | [34] |
Table 2. A summary of the various detection methods for single EVs
| Detection methods | Working principle | Information detected | Advantages | Disadvantages | Ref. |
|
| Label-free approaches | RTM | Refer to Table 1 | [39] |
|
| Surface-enhanced Raman spectroscopy (SERS) | Exposing EVs to signal-enhancing nanoparticles to obtain amplified Raman signal | SERS spectra of individual EVs | Enhanced Raman signal, high sensitivity, high throughput | Limited information of EV surface proteins, low reproducibility, complicated data processing | [43, 44] |
|
| Surface plasmon resonance imaging (SPRi) | A technique which detects the changes in refractive index induced by molecular binding to noble metal surface | SPR spectra and SPR imaging upon binding of EVs on substrate | High sensitivity, multiplexed data collection, compatibility with microfluidic systems | Low throughput, uniform substrate and modified sensor surface required | [45, 46] |
|
| Interferometric plasmonic microscopy (iPM) | Combined surface plasmon stimulation and interferometric scattering effect to detect single EVs | iPM images reflecting the adsorption and binding events of single exosomes | High sensitivity, high spatial resolution, in situ visualization | Low specificity, low throughput | [47] |
|
| Single-particle interferometric reflectance imaging sensor (SP-IRIS) | Detecting individual enhanced scattering signals generated by bound vesicles on layered substrate | Size and multiplexed profiling of membrane biomarkers of EV groups in a single measurement | High sensitivity, high specificity, high throughput, compatibility with microfluidics | Limited lateral resolution, difficult to detect small nanovesicles | [18, 48] |
|
| Frequency locking optical whispering evanescent resonator (FLOWER) | Recognizing EVs by tracking changes in resonant frequency of the microtoroid optical resonators | Resonance frequency changes over time indicating the binding of EVs to resonator | High signal-to-noise ratio, high sensitivity | Difficult to identify particle size, limited information obtained | [49] |
|
| Reflection enhanced dark field scattering microscopy (REDFSM) | Recording scattering signal of single EVs after illumination on a reflective surface | The size and scattering intensity of single exosomes | High sensitivity, simplicity, high spatial resolution | Low throughput, low specificity | [50] |
|
| Labelling approaches | Imaging flow cytometry (IFCM) | Detection by combined flow cytometry and imaging techniques | Fluorescence images for qualitative and quantitative evaluation of single EVs | Multi-parametric detection, high throughput, stable signal | Dependence on biological reference materials, time consuming | [51, 52] |
|
| Micro/ Nano-Flow cytometry (MFC/nFC) | Refer to Table 1 | [53-55] |
| Detection methods (continued) | Working principle | Information detected | Advantages | Disadvantages | Ref. |
|
| Fluorescence microscopic imaging | Fluorescence from external labels for in vitro and in vivo tracking | Fluorescence images of single EVs both in vitro and in vivo | Rapid response, multi-colour labelling, high sensitivity, compatibility with microfluidics | Low signal-to-noise ratio, fluctuation of fluorescence induced by low photostability of dyes | [56-61] |
|
| Super-resolution microscopy (SRM) | Fluorescence from external labels detected below the optical diffraction limit | Fluorescence images of single EVs in biological samples | High resolution, in situ visualization, less invasive | Fixed samples required, interference from fixing regent, low throughput | [62] |
|
| Total internal reflection fluorescence (TIRF) microscopy | Imaging of fluorescence signal generated by the evanescent wave induced excitation of external labels at glass-water interface | Fluorescence images of single EVs in aqueous environment | High axial resolution, high signal-to-noise ratio | Limited specimen detected, fluorophore instability and photobleaching | [63, 64] |
|
| DNA points accumulation for imaging in nanoscale topography (DNA-PAINT) | Combination of DNA-PAINT and TIRF to measure exosomal biomarkers | Quantitative analysis of exosomal surface biomarkers on individual vesicles | High accuracy, high resolution, multiplexed profiling | Complicated data analysis | [65] |
|
| Droplet digital exosome enzyme-linked immunosorbent assay (ExoELISA) | Detection of the enzymatic fluorescent reporter tagged on sandwich ELISA complexes of exosomes in single droplets | Fluorescence signals from droplets indicating the expression levels of biomarker on single exosomes | High specificity, high sensitivity, high accuracy, high throughput | Magnetic beads dependence, diluted sample required | [66] |
|
| Immune droplet digital polymerase chain reaction (iddPCR) | PCR amplification of the genetic barcode of labelled EVs in single droplets | Absolute quantification of specific targets in individual EVs | Multiplexed analysis, high sensitivity, high throughput | Difficulty in obtaining absolute expression level of biomarker, diluted sample required | [67] |
|
| Proximity barcoding assay (PBA) | Profiling surface proteins of individual EVs using antibody-DNA conjugates and next-generation sequencing | Surface protein patterns of individual EVs | Multiple-recognition assay, high throughput, high specificity | Complicated processing, time consuming | [68] |
|
| Nanoplasmon-enhanced scattering (nPES) | Local plasmon-coupling effect induced by the binding of Au nanostructures to immunocaptured EVs | Dark field images and scattering spectra indicating the concentration of EV and biomarker | High specificity, high sensitivity, no sample pre-treatment, little sample required | Low throughput | [46] |
|
| Nano-plasmonic EV analysis with enhanced fluorescence detection (nPLEX-FL) | Fluorescence signals of the immunostained EVs are amplified by SPR excited by the Au nanohole structures | Enhanced fluorescence intensity indicating the biomarker profiling of single EVs | Multiplexed analysis, high sensitivity | Complex substrate modification, limited enhancement of fluorescence | [69] |
|
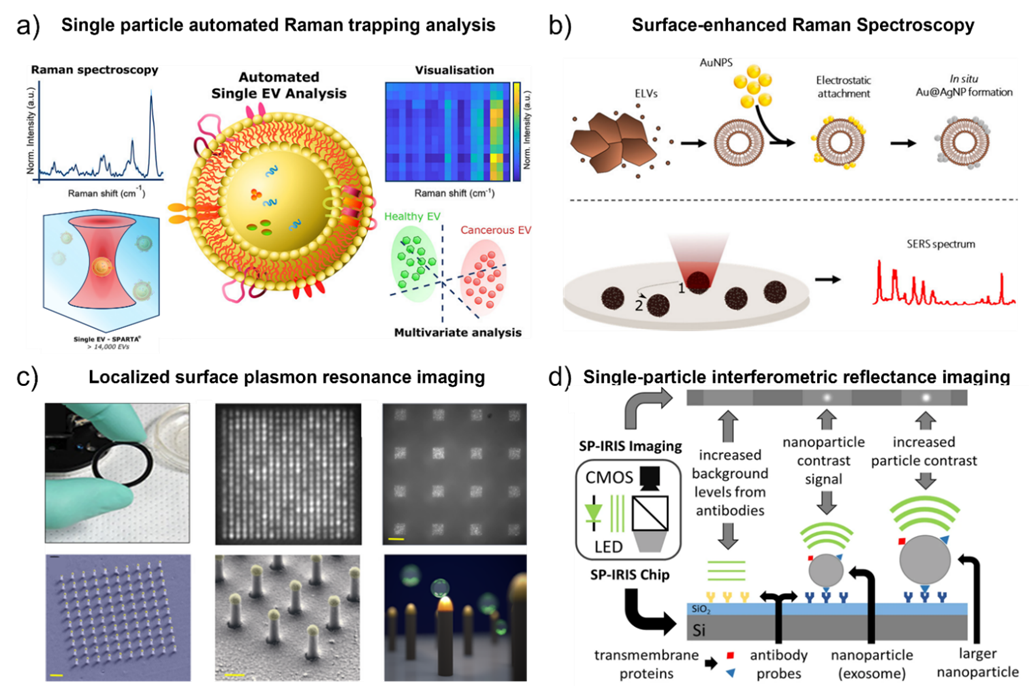
Fig. 2. a) Schematic illustration of the automated single EV analysis using the Single Particle Automated Raman Trapping Analysis (SPARTA). Adapted with permission from ref. [39] Copyright 2021 American Chemical Society. b) Schematic illustrateion of the label-free identification of individual exosome-like vesicles with Au@Ag nanoparticles as SERS substrate. Adapted with permission from ref. [44] Copyright 2019 American Chemical Society. c) Schematic diagram of the localized surface plasmon resonance imaging platform. Adapted with permission from ref. [72]. Copyright 2018 PLoS one. d) Detection principle of single-particle interferometric reflectance imaging (SP-IRIS). Adapted with permission from ref. [48]. Copyright 2016 Springer Nature.
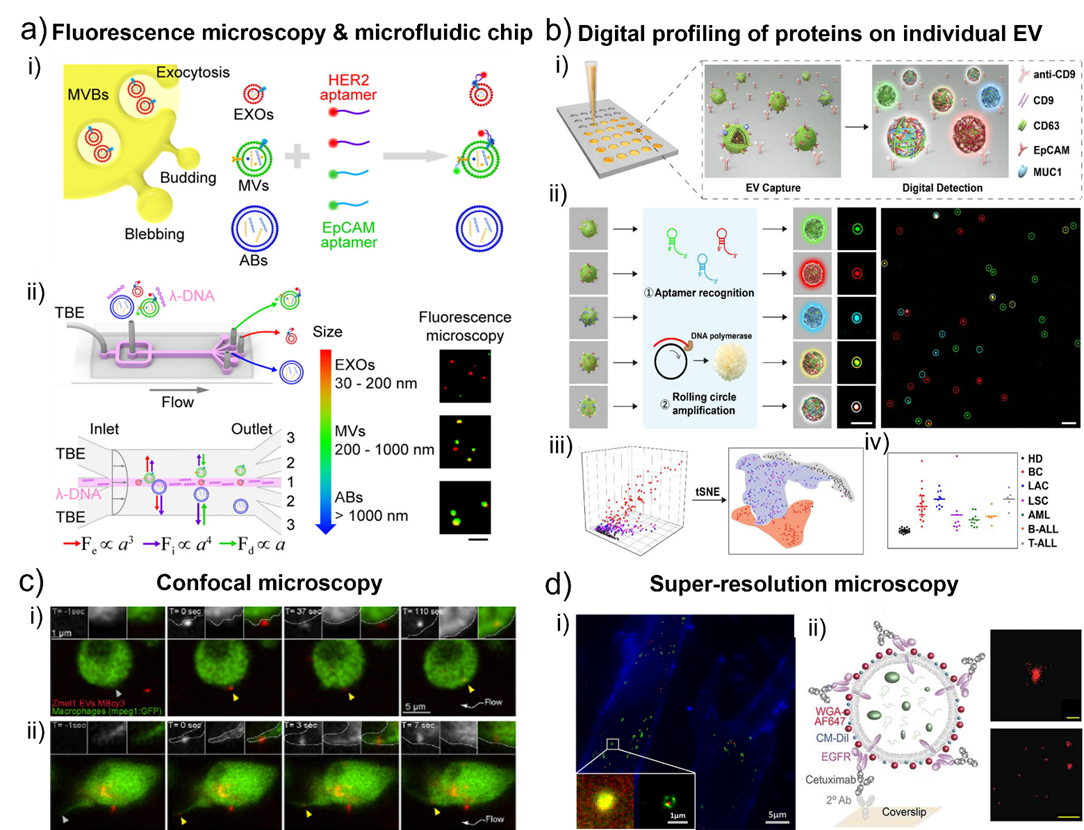
Fig. 3. a) Schematic of l-DNA-mediated sorting of EV subpopulations and aptamer-based analysis of individual EVs. i) Labeling of cell originating EVs including exosomes (EXOs, red), microvesicles (MVs, green), and apoptotic bodies (ABs, blue) with fluorescent HER2 and EpCAM aptamers. ii) Size-selective separation of EV subpopulations. Fluorescence microscopy images showed HER2 (red) and EpCAM (green) expression of isolated individual EVs. Scale bar, 5 μm. Adapted with permission from ref. [58]. Copyright 2018 American Chemical Society. b) Schematic diagram of digital profiling of proteins on individual EV (DPPIE) for high-dimensional individual EV analysis and multi-cancer classification. i) Overview of the steps in DPPIE assay. ii) The captured EV is labelled with DNA aptamers, followed by RCA to generate localized amplified fluorescent signals that can be imaged by confocal microscopy. Fluorescence images showed CD63 (green), EpCAM (red) and MUC1 (blue) expression on individual EV. Scale bar: 3 μm. iii) Individual EV analysis data from different cells are classified using tSNE algorithm. iv) Multi-cancer diagnosis and classification. Adapted with permission from ref. [57]. Copyright 2020 John Wiley and Sons. c) Single-plane confocal images of Tg (mpeg1: GFP) embryos injected with Zmel1 MemBright-Cy3 (MBCy3) EVs extracted from time-lapses generated immediately after injection and showing: i) the attachment and uptake of EVs by endocytosis and ii) the sliding of EVs on the macrophage protrusion and its fast internalization. Adapted with permission from ref. [60]. Copyright 2019 Elsevier. d) i) Colocalization of MRC-5 lysosomes (green) and internalized SKBR3 exosomes (red). Figure inset showed the enlarged dual-color PALM/STORM image that exosomes were clearly visualized in the interior of lysosomes. Adapted with permission from ref. [65]. Copyright 2016 American Chemical Society. ii) Left, scheme of an EV affinity isolated with cetuximab and labelled with WGA-AF647 as a reporter. Right, filtered dSTORM images of WGA-AF647 with localizations in red. A single EV (top; scale bar, 100 nm) and a larger field of view (bottom; scale bar, 1 µm) are shown. Adapted with permission from ref. [79]. Copyright 2019 Taylor & Francis Group.
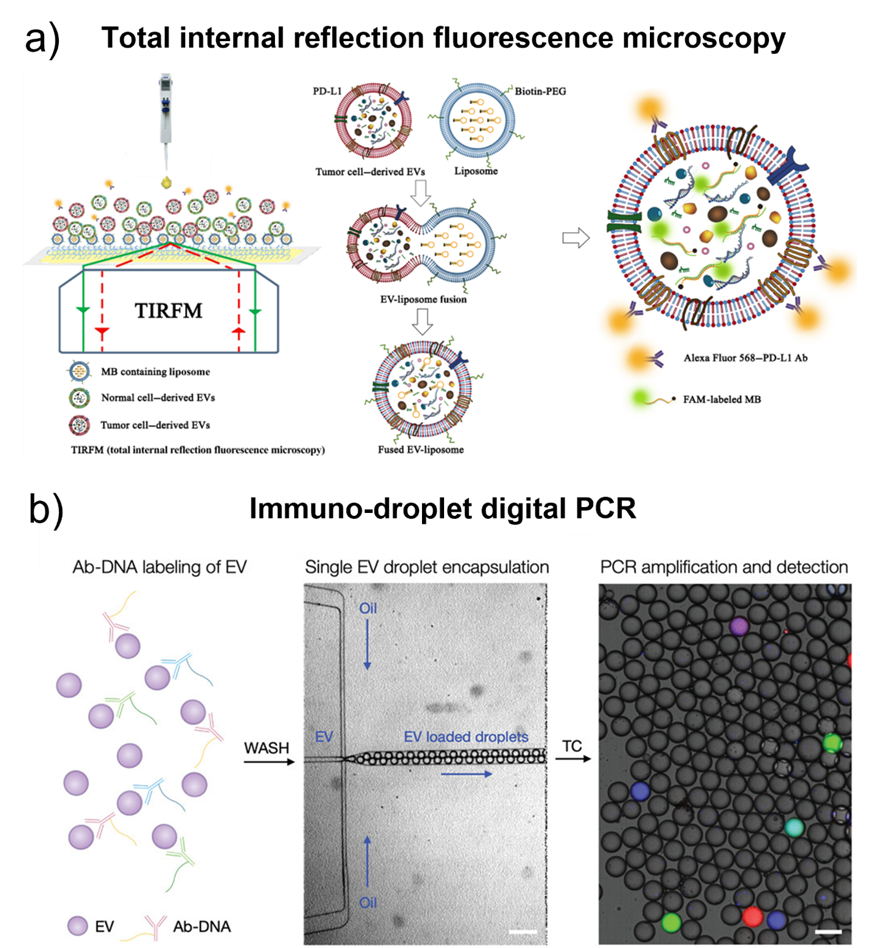
Fig. 4. a) Illustration of the HNCIB system for simultaneous detection of PD-L1 membrane protein and mRNA in a single EV. Adapted with permission from ref. [80]. Copyright 2020 American Association for the Advancement of Science. b) Schematic of droplet-based single EV detection. Adapted with permission from ref. [67]. Copyright 2020 John Wiley and Sons.
Table 3. Summary of the main application based on single EV detection for disease diagnosis
| Disease type | Sample | Biomarkers | Patient cohort/ healthy control (HC) | Sample volume | Isolation technique | Detection method | Analysis time | Diagnostic accuracy (%) | Ref. |
| Breast cancer | Serum | CD47 | Cancer patients n = 60/ HC n = 60 | 500 µL | Ultracentrifugation | Micro flow cytometry (MFC) | ~55 min | N/A | [53] |
| Serum | GPC-1 | Cancer patients n=12/ patients with benign disease n=5/ patients after surgery n=2/ HC n=5 | 50 μL | Ultracentrifugation | Droplet digital ExoELISA | ~2 h | ~100 | [66] |
| Serum | HER2, EpCAM | Cancer patients n = 7/ HC n = 4 | 2.5 μL | Differential centrifugation | Fluorescence microscopy | >40 min | 97.42 | [58] |
| Plasma | CD63, EpCAM, MUC1 | Cancer patients n = 14/ HC n=15 | 1 μL | Immunocapture on biochip | Digital Profiling of Proteins on Individual EV (DPPIE) assay | >1h15min | 91 | [57] |
| Lung cancer | Plasma | miR-21, PD-L1 | Cancer patients n = 34 /HC n = 35 | ~90 µL | Precipitation & centrifugation | High-throughput Nano-bioChip Integrated System for Liquid Biopsy (HNCIB) assay | ~6 h | N/A | [80] |
| Serum | PD-1/PD-L1 mRNAs | Cancer patients n = 27/ HC n = 27 | 3 µL | Filtration & centrifugation | High-resolution TIRF microscopy on AuSERP biochip | ~ 2 h | 93.2 | [89] |
| Plasma | CD63, EpCAM, MUC1 | Cancer patients of subtype I n = 10/ Cancer patients of subtype II n = 9/ HC n=15 | 30 μL | Immunocapture on biochip | DPPIE assay | >1h15min | N/A | [57] |
| Pancreatic cancer | Plasma | EphA2 | Cancer patients n = 27/ HC n = 27 | 1 µL | Immunocapture on nanoplasmon-enhanced scattering (nPES) platform | Dark-field microscopy on nPES platform | 5 h | 94 | [46] |
| Plasma | EGFR, CA19-9 | Cancer patients n = 5/ HC n = 6 | ~200 μL | Size exclusion chromatography | Quantitative single molecule localization microscopy (qSMLM) assay | ~2.5 h | N/A | [79] |
| Serum | glypican 1 | Cancer patients n = 56/ patients with benign disease n = 6/ HC n = 20 | 250 μL | Ultracentrifugation | Flow cytometry | >2.5 h | 100 | [90] |
| Serum | HULC lncRNA | Cancer patients n = 20/ patients with benign disease n = 22/ HC n = 21 | 500 μL | Ultracentrifugation | Digital PCR | >30 min | 92 | [91] |
| Colorectal cancer | Plasma | CD147 | Cancer patients n = 37/ HC n = 32 | 50 μL | Ultracentrifugation | High-sensitivity flow cytometry (HSFCM) | ~7 h | 93.2 | [55] |
| Plasma | Somatic BRAF and KRAS mutations | Cancer patients n = 21/ HC n =46 | 1 mL | Nickel-based isolation | Droplet digital PCR | N/A | 93 | [92] |
| Prostate cancer | Plasma | Circulating prostate microparticles (PMPs) | Cancer patients n = 256/ patients with metastatic cancer n=67/ patients with benign disease n = 156/ HC n = 22 | 20 μL | Fluorescence-activated cell sorting (FACS) isolation | Nanoscale flow cytometry | N/A | N/A | [93] |
| Serum and plasma | Beta-sheet-rich proteins | Cancer patients n = 4/ patients with benign disease n = 4/ HC n = 1 | 2 mL | Differential centrifugation | Infrared & Raman spectroscopy | > 3 h | N/A | [94] |
| Plasma | AR-V7 mRNA | Cancer patients n=16/patients with metastatic cancer n = 35/ HC n=10 | 500 μL | Ultracentrifugation | Droplet digital PCR | > 2 h | N/A | [95] |
| Serum | PSA mRNA | Cancer patients n=27/ patients with benign prostatic hyperplasia n=15/ HC n=16 | 2 μL | FRET-based DNA tetrahedron (FDT) incubation | DNA tetrahedron-based thermophoretic assay & qRT-PCR | ~2h45min | 0.93 | [96] |
| Glioma cancer | Cerebrospinal fluid | Mutant IDH1 transcript | Cancer patients n=14/ HC n=4 | 500 μL | Ultracentrifugation | BEAMing RT-PCR & droplet digital PCR | ~ 2h | N/A | [97] |
| Leukaemia | Plasma | CD63, EpCAM, MUC1 | Cancer patients with subtype I n = 10/ patients with subtype II n=5/ patients with subtype II n=5/ HC n=15 | 30 μL | Immunocapture on biochip | DPPIE assay | >1h15 min | N/A | [57] |
| Plasma | Tyrosine protein kinase-like 7 (PTK7) | Cancer patients n=5/ HC n=5 | 50 μL | Immunocapture on coverslip | TIRF assay | >7 h | N/A | [63] |
Fig. 6. Overview of critical future perspectives in single EVs analysis and related biomedical applications.
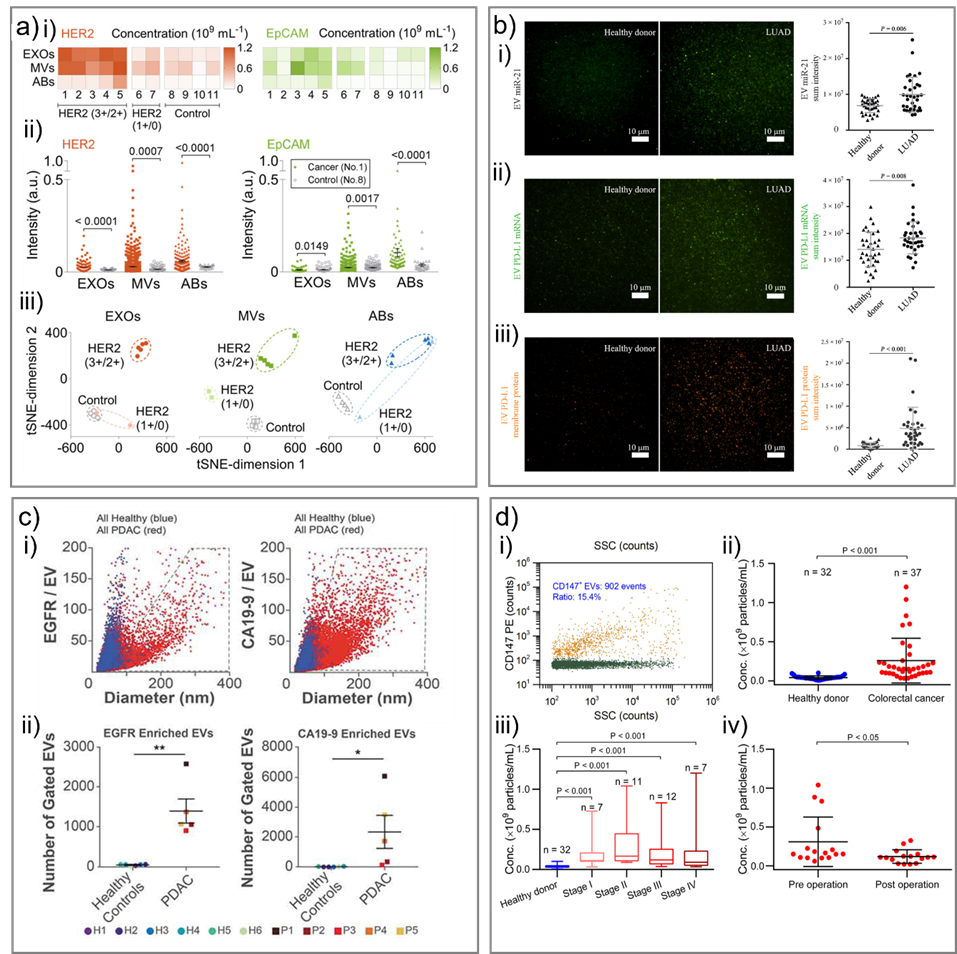
Fig. 5. Representative studies in cancer diagnostics using single EV analysis. a) Discrimination of the Stage II breast cancer patients and healthy controls. i) The concentrations of HER2-positive EXOs, MVs, and ABs in the clinical cohort. ii) Comparison of expression levels of HER2 and EpCAM on individual EXOs, MVs, and ABs between a cancer patient and a control. iii) t-SNE visualization of discrimination among cancer patients with varied HER2 expressions and controls by different EV subpopulations. Adapted with permission from ref. [58]. Copyright 2019 American Chemical Society. b) Measurement of miRNA, mRNA, and protein expression of EVs isolated from human plasma. Samples were from 35 healthy donors and 34 patients with LUAD. Representative images and statistical analysis of i) EV miR-21, ii) EV PD-L1 mRNA and iii) EV PD-L1 membrane protein. Adapted with permission from ref. [80]. Copyright 2020 American Association for the Advancement of Science. c) Quantification of EVs from patient plasma. i) Combined EGFR-enriched (left) and CA19-9-enriched (right) EVs from healthy subjects and PDAC patients. ii) Number of EVs in gated area (grey polygons, panel i). Adapted with permission from ref. [79]. Copyright 2019 Taylor & Francis Group. d) Analysis of CD147-positive EVs in plasma samples of colorectal cancer patients and heathy donors by HSFCM. i) Bivariate dot-plot of the PE orange fluorescence versus SSC for EVs from patient sample upon immunofluorescent staining with PE-conjugated MAb against CD147. Comparison of plasma particle concentrations of CD147-positive EVs in ii) healthy donors and colorectal cancer patients, iii) healthy donors and cancer patients at different stages and iv) cancer patients (n = 16) before (preoperation) and after (7–10 days postoperation) surgical removal of the tumor (mean ± s.d.). Adapted with permission from ref. [55]. Copyright 2018 American Chemical Society.
Fig. 6. Overview of critical future perspectives in single EVs analysis and related biomedical applications.
6.Conclusions and future perspectives
EVs are present in nearly all types of biofluids and contain membrane and cytoplasmic components representing the parental cells, including proteins, nucleic acids, and lipids. Findings on the role of EVs have indicated their diverse biological functions, which makes EVs be reliable biomarkers for diseases diagnosis. Increasing evidence has demonstrated that EVs are heterogeneous in size and molecular contents, which accordingly change their biological properties. This intrinsic heterogeneity introduces extra complexity in studying EV biology and function. However, reported methods focus mainly on the bulk analysis of vesicles, which makes it challengeable to study the various EV types and subpopulations, as well as understanding the distinct biomarker expression. Large efforts have been devoted to overcoming these challenges and producing single EV-optimized analytical platforms. Most of novel analytical technologies are superior to traditional methods, effectively measuring the specific properties from single vesicle. In this article, as summarized in Table 2, current and emerging approaches for analysis of single EVs are categorized into label-free and labelling groups. Label-free methods including RTM, SERS and SPR etc. are based on the analysis of EVs’ physical characteristics and overall spectral properties. The labelling methods are mainly based on modifying surface protein with fluorescent dyes, in combination with FC, TIRF or high-resolution imaging techniques to reveal the detection of individual vesicles. Technologies like microfluidics has been successfully integrated with various labelling approaches, achieving the detection of EVs at single vesicle level.
The development of single EVs analysis accelerates not only the understanding of EVs’ biology, but also the improved biomedical application with prominent disease diagnosis potential. With an emphasis for cancer investigations, we conclude several critical points including low volume of body fluid to analyse EVs, rapid isolation and analysis of single EVs, reliable biomarkers for cancer diagnosis, high throughput EV analysis and high diagnostic accuracy etc. These technological parameters are crucial for the evaluation of analysis techniques, with the aim to promote the translation to clinics.
Despite the increasing knowledge of EV subpopulations after decades, EV-based research is still in its infancy. Limitations in isolation and detection of single EVs, as well as the in-depth understanding of the roles of EV subpopulations in physiology remain hurdles in the development of EVs research. Based on the advances and the remaining challenges in single EV-related research, we describe the critical issues that we consider for future development in this field (Fig. 6). First, advanced single EVs isolation technologies are still demanded to investigate the exosome-to-exosome phenotypic heterogeneity and their accurate quantification. Owing to the nanometer sizes, isolation of single EVs is still challengeable. In this regard, microfluidic and nanofluidic devices show their advantages due to the comparable dimensions. Combined with nanopatterns and immune-affinity technology, the microfluidic systems are expected to isolate single EVs with high sensitivity and specificity. Therefore, more innovative microfluidic chips with elaborate design are highly desired to achieve highly efficient isolation of single EVs, not only from cell culture medium, but also from complicated clinical samples.
Second, single EVs research requires highly sensitive detection to address the heterogeneity of biophysical characteristics and composition of EVs. As a result, advanced techniques such as fluorescent imaging-based high-resolution microscopy is under focus. Super resolution microscopy has been utilized for imaging of single EVs, due to the ultrahigh resolution down to a few tens of nanometers. Research efforts has been devoted to the development of instrumentation and labelling methods to realize highly sensitive single EV detection. Therefore, in one aspect, the advancement of instrument, for instance three-dimensional super-resolution microscopy will be of utmost importance for investigating the unique characteristics including sizing and biomarker expression of single nanovesicles. In another aspect, promising labelling strategies such as immunoaffinity combined with DNA assisted extension will help to amplify the signals, which enables sensitive quantification of the surface proteins of single EVs. Furthermore, microfluidic systems provide an advanced platform for single EVs detection. Taking advantage of the nanochannels and precise fluid control, individual vesicles can be visualized after proper fluorescence labelling.
Third, the development of single EV research will be based on the joint efforts of associated technologies. And thus, multidisciplinary research including nanotechnology, fluorescence imaging, micro-fabrication and machine learning etc. will contribute to the advancing of single EV-related field. For instance, further advancement in micro- and nanofabrication will offer powerful platforms for studying the single EV transportation by mimicking biosystems (e.g., tumor microenvironment).
After combining with functional labelling linkers and advanced imaging techniques, precise investigation on the roles of EV subpopulation in disease development would accelerate the single EV-based research.
Finally, biomedical application of single EV analysis still relies on the advancement of powerful detection approaches and more clinical practice to generate translational effect. In the past few years, precise medicine, such as personalized pharmacotherapy and tumor screening, has gained great attention in clinical oncology. The increasing development of EV-related research provides tremendous prospects for biomedicine. Biomedical applications based on single EV strategies require the merits of low sample consumption, rapid response, early detection, and capability of differentiation between different stages of certain cancer types. Therefore, the various emerging systems for detecting individual EVs will be beneficial to understanding the role of EV subpopulations in cancer biology and facilitating the development of new strategies for clinical diagnosis and therapeutics. For cancer diagnosis, since different EV subpopulations exert unique roles in tumor development, the screen of reliable biomarkers via single EV analysis has great potential to facilitate accurate cancer diagnosis. Additionally, cell derived EVs have been regarded to be promising drug carriers, due to their nanoscale structure, biocompatibility, and their capability in crossing blood-brain barrier. Therefore, EV heterogeneity must be considered in terms of biological distribution and intracellular delivery, and specific EV subtypes could be emerging efficient drug delivery tools for precision therapeutics.

